Detecting Mesoderm Invagination¶
Calculate the time when mesoderm invagination occurs in order to align samples in time and to start point tracking at the start of germband invagination
import numpy as np
import pandas as pd
import matplotlib
matplotlib.use('agg')
import matplotlib.pyplot as plt
import tifffile
import av
import tqdm
import gbeflow
fs = ['20180108_htl_glc_sc11_mmzm_rotate_brt',
'20180108_htl_glc_sc2_mmzm_wp_rotate_brt',
'20180108_htl_glc_sc9_mmzp_rotate_brt',
'20180110_htl_glc-CreateImageSubset-01_sc10_wt_rotate_brt',
'20180110_htl_glc-CreateImageSubset-02_sc11_htl_rotate_brt',
'20180110_htl_glc_sc14_mmzp_rotate_brt',
'20180110_htl_glc_sc15_mmzm_rotate_brt',
'20180110_htl_glc_sc6_mmzm_rotate_brt',
'20180112_htlglc_tl_sc11_mmzp_rotate_brt',
'20180112_htlglc_tl_sc4_resille_rotate_brt']
Select a single sample as a test case
f = '20180110_htl_glc_sc14_mmzp_rotate_brt'
Load vector data and movie mesoderm_figs/output
# vimg = gbeflow.load_avi_as_array(f+'.avi')
vf = gbeflow.VectorField(f)
expected = 35
fig,ax = plt.subplots(figsize=(10,8))
ax.imshow(vimg[expected])
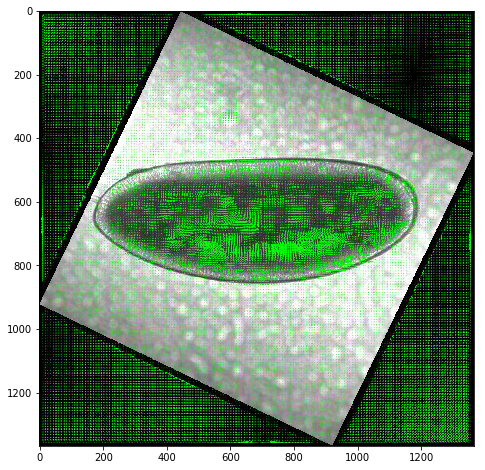
Test metrics for detecting mesoderm invagination¶
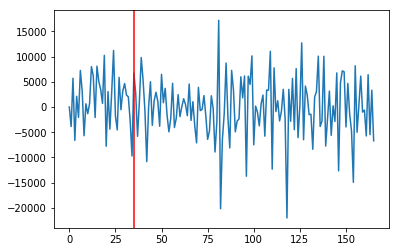
Sum of y component of vectors over time¶
vf.vy.shape
(166, 270, 270)
ysum = np.sum(vf.vy,axis=(1,2))
fig,ax = plt.subplots()
ax.plot(ysum)
ax.axvline(expected,c='r')
<matplotlib.lines.Line2D at 0x109358d30>
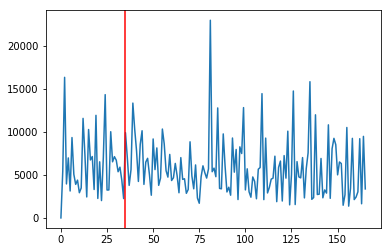
Try looking at the sum of only positive vy components¶
vfpos = np.copy(vf.vy)
vfpos[vfpos<0] = 0
ysum = np.sum(vfpos,axis=(1,2))
fig,ax = plt.subplots()
ax.plot(ysum)
ax.axvline(expected,c='r')
<matplotlib.lines.Line2D at 0x1d21e1fc50>
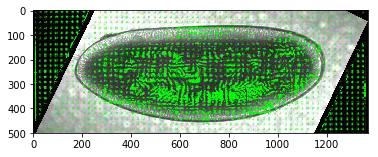
What about within an roi¶
roi = np.s_[:,400:900,:]
fig,ax = plt.subplots()
ax.imshow(vimg[roi][35])
<matplotlib.image.AxesImage at 0x11579f0b8>
np.interp(a, (a.min(), a.max()), (-1, +1))
roirange = np.interp([400,900], (0,1368), (0,270))
roirange
array([ 78.94736842, 177.63157895])
vroi = np.s_[:,int(roirange[0]):int(roirange[1]),:]
vfpos[vroi].shape
(166, 99, 270)
ysum = np.sum(vfpos[vroi],axis=(1,2))
fig,ax = plt.subplots()
ax.plot(ysum)
ax.axvline(expected,c='r')
<matplotlib.lines.Line2D at 0x1d239af9e8>
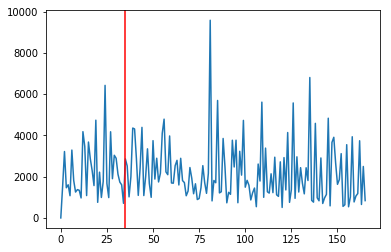
Sum of the squared positive vy¶
ysum = np.sum(np.power(vfpos[vroi],2),axis=(1,2))
fig,ax = plt.subplots()
ax.plot(ysum)
ax.axvline(expected,c='r')
ax.axvline(55,c='m')
<matplotlib.lines.Line2D at 0x1d24b19eb8>
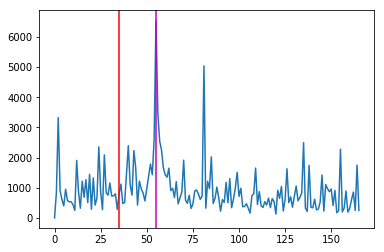
fig,ax = plt.subplots(figsize=(10,8))
ax.imshow(vimg[roi][55])
<matplotlib.image.AxesImage at 0x1d24c678d0>
While we are not detecting mesoderm invagination here, this feature does mark germband extension which could be useful in itself.
Let’s try this on other samples to see if the feature is consistent¶
Dvimg = {}
for f in fs:
try:
Dvimg[f] = gbeflow.load_avi_as_array(f+'.avi')
except:
print('Video import failed',f)
Dvf = {}
for f in Dvimg.keys():
try:
Dvf[f] = gbeflow.VectorField(f)
except:
print('Import failed',f)
Dsum = {}
for f in Dvimg.keys():
vfpos = np.copy(Dvf[f].vy)
vfpos[vfpos<0] = 0
Dsum[f] = np.sum(np.power(vfpos[vroi],2),axis=(1,2))
for f in Dsum.keys():
fig,ax = plt.subplots(1,2,figsize=(10,8))
ax[0].plot(Dsum[f])
ax[0].axvline(maxsum[f],c='r')
ax[0].set_title(f)
ax[1].imshow(Dvimg[f][roi][maxsum[f]])
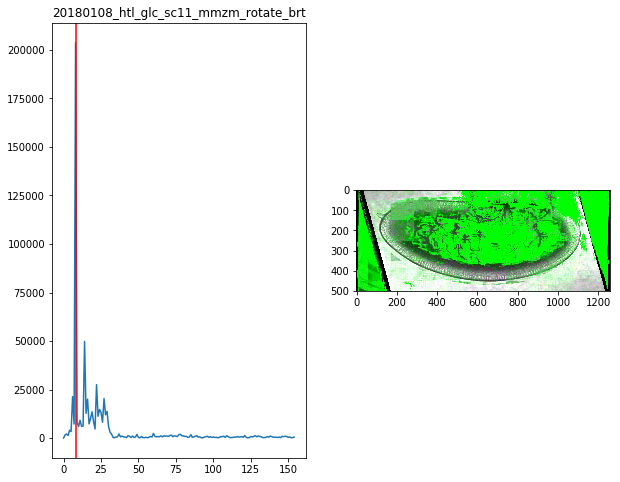
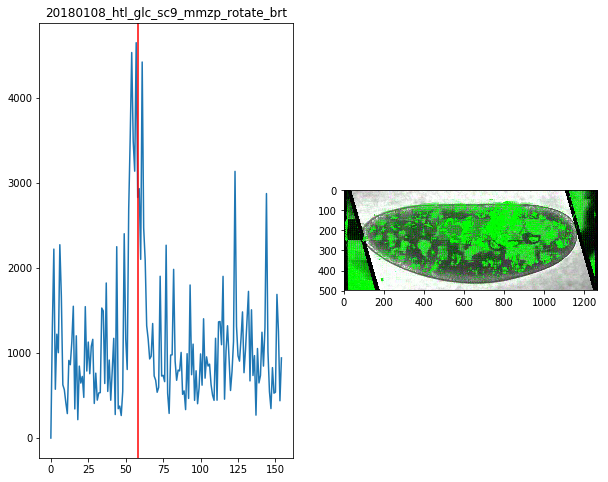
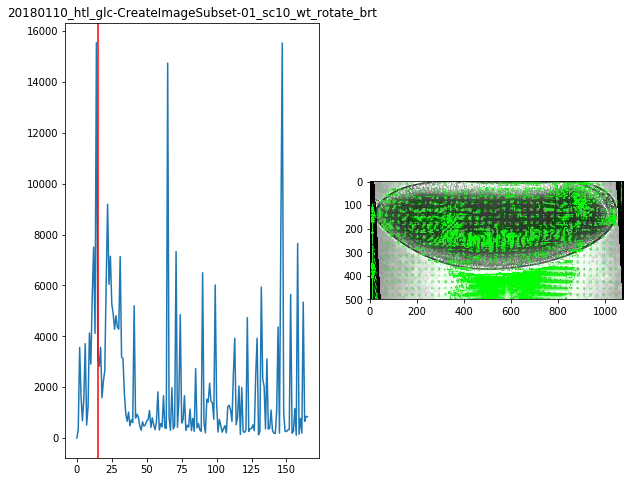
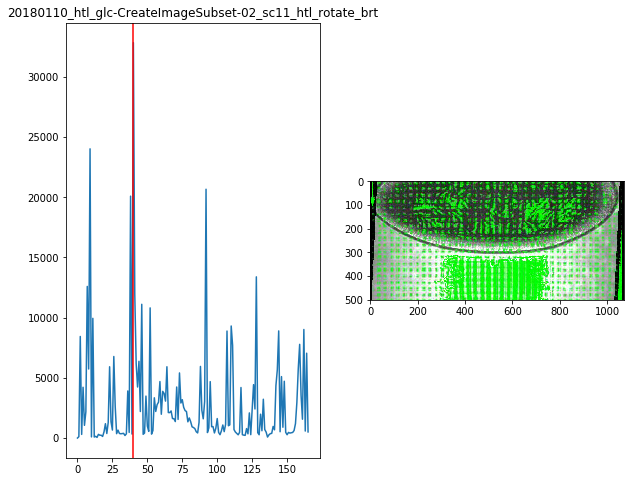
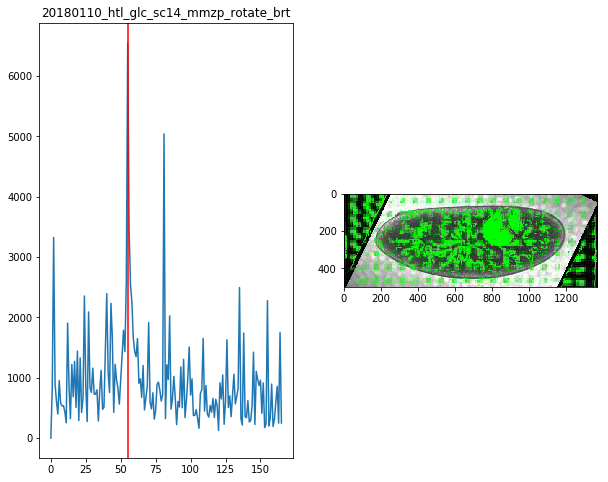
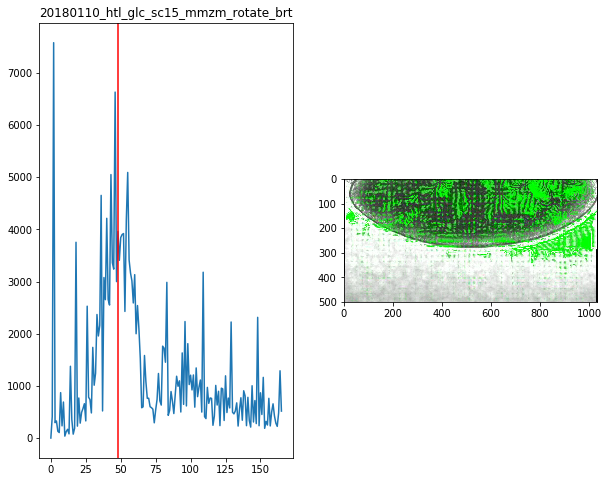
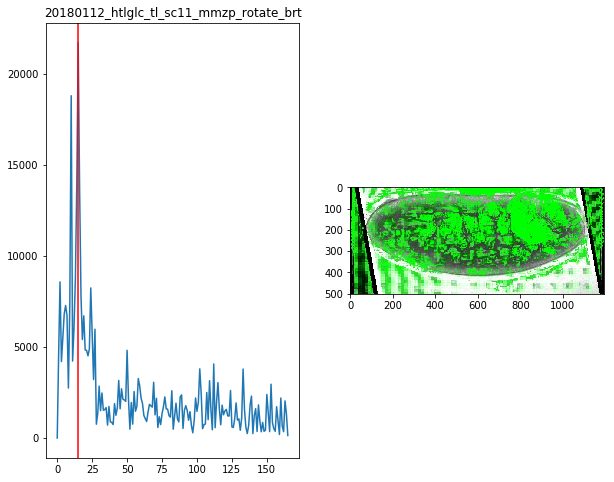
maxsum = {
'20180108_htl_glc_sc11_mmzm_rotate_brt':8,
'20180108_htl_glc_sc9_mmzp_rotate_brt':58,
'20180110_htl_glc-CreateImageSubset-01_sc10_wt_rotate_brt':15,
'20180110_htl_glc-CreateImageSubset-02_sc11_htl_rotate_brt':40,
'20180110_htl_glc_sc14_mmzp_rotate_brt':55,
'20180110_htl_glc_sc15_mmzm_rotate_brt':48,
'20180112_htlglc_tl_sc11_mmzp_rotate_brt':15
}
For the time being it looks like manually selecting the timepoint of mesoderm invagination will be more expedient.
Check manual assignments¶
tpoints = pd.read_csv('mesoderm_invagination.csv')
tpoints
| File | Timepoint | |
|---|---|---|
| 0 | 20180110_htl_glc_sc14_mmzp_rotate_brt.avi | 36 |
| 1 | 20180108_htl_glc_sc9_mmzp_rotate_brt.avi | 36 |
| 2 | 20180108_htl_glc_sc11_mmzm_rotate_brt.avi | 62 |
| 3 | 20180110_htl_glc_sc15_mmzm_rotate_brt.avi | 41 |
| 4 | 20180110_htl_glc-CreateImageSubset-01_sc10_wt_... | 16 |
| 5 | 20180110_htl_glc-CreateImageSubset-02_sc11_htl... | 57 |
| 6 | 20180112_htlglc_tl_sc11_mmzp_rotate_brt.avi | 4 |
tpoints[tpoints['File']==f+'.avi'].values[-1,-1]
62
for f in Dvimg.keys():
fig,ax = plt.subplots(figsize=(10,8))
t = tpoints[tpoints['File']==f+'.avi'].values[-1,-1]
ax.imshow(Dvimg[f][t])
ax.set_title(f)
Test track interpolation starting at mesoderm invagination¶
tracks = pd.read_csv('20181128-tracking.csv')
tracks.head()
starts = tracks[tracks.t==0][['f','x','y']]
trange = range(maxsum[f],np.max(vf.tval))
(trange)
xpos = [x0]*(maxsum[f]+1)
ypos = [y0]*(maxsum[f]+1)
for t in trange:
dx = vf.Ldx[t].ev(xpos[t],ypos[t])
dy = vf.Ldy[t].ev(xpos[t],ypos[t])
xpos.append(xpos[t] + dx*60)
ypos.append(ypos[t] + dy*60)
track = np.array([xpos,ypos])
trackdf = pd.DataFrame({'x':track[0,:],'y':track[1,:],'t':vf.tval,
'track':[i]*track.shape[-1],
'name':['test']*track.shape[-1]})
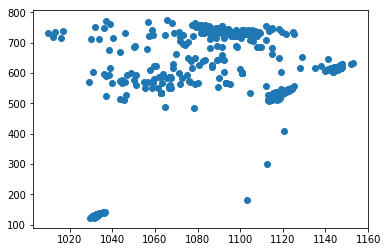
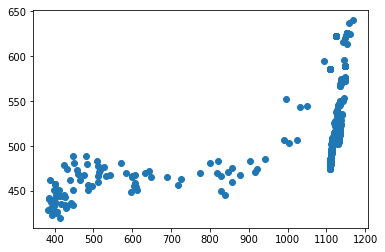
Track visualization¶
tracks = pd.concat(Ltrack,keys=list(Dvf.keys())
).reset_index(
).drop(columns=['level_1']
).rename(columns={'level_0':'file'})
for f in tracks['file'].unique():
fig,ax = plt.subplots()
sb = tracks[tracks['file']==f]
ax.scatter(sb.x,sb.y)#,c=sb.t,cmap='plasma')
for f in tracks['file'].unique():
gbeflow.make_track_movie(Dvimg[f],tracks[tracks.file==f],c='r',
name='20181202_'+f+'_tracks')
Attempting to start interpolating the tracks at the time of mesoderm invagination does not appear to improve the results of the tracking since there is still a large fraction that wanders out of the frame entirely.